How to use reval¶
In the following, we are going to simulate N = 1,000 sample dataset with two groups and two features
(for visualization purposes), then we will show how to apply the reval package and investigate
the result types. We will use hierarchical clustering and KNN classification algorithms.
First (after starting ipython or a jupyter notebook),
let us import a bunch of useful libraries and our class reval.best_nclust_cv.FindBestClustCV:
from reval.best_nclust_cv import FindBestClustCV
from sklearn.datasets import make_blobs
from sklearn.neighbors import KNeighborsClassifier
from sklearn.cluster import AgglomerativeClustering
from sklearn.model_selection import train_test_split
import matplotlib.pyplot as plt
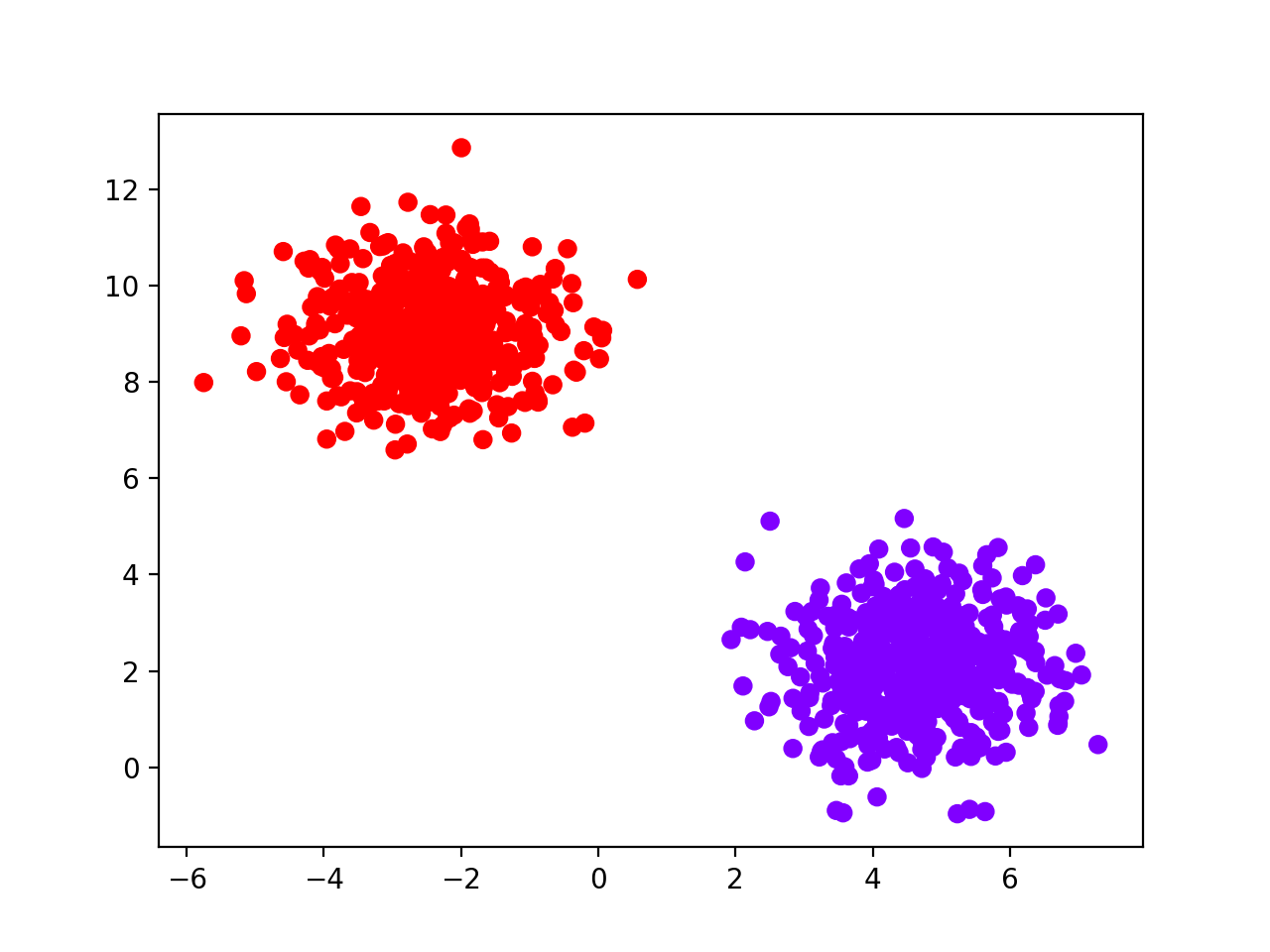
Then we simulate the toy dataset and visualize it:
data = make_blobs(1000, 2, centers=2, random_state=42)
plt.scatter(data[0][:, 0], data[0][:, 1],
c=data[1], cmap='rainbow_r')
plt.show()
Then, we split the dataset into training and test sets:
X_tr, X_ts, y_tr, y_ts = train_test_split(data[0], data[1],
test_size=0.30,
random_state=42,
stratify=data[1])
We apply the stability-based relative clustering validation approach with 10x2 repeated cross-validation, 10 iterations of random labeling, and number of clusters ranging from 2 to 10.
classifier = KNeighborsClassifier()
clustering = AgglomerativeClustering()
findbestclust = FindBestClustCV(nfold=2,
nclust_range=list(range(2, 11)),
s=classifier,
c=clustering,
nrand=100)
metrics, nbest = findbestclust.best_nclust(X_tr, iter_cv=10, strat_vect=y_tr)
out = findbestclust.evaluate(X_tr, X_ts, nbest)
To obtain the training stability and the normalized validation stability for the selected number of clusters we need to call:
nbest
# 2
metrics['train'][nbest]
# (0.0, (0.0, 0.0)) (stab, (stab, error))
metrics['val'][nbest]
# (0.0, (0.0, 0.0)) (stab, (stab, error))
out returns train/test accuracies and test set clustering labels.
out.train_cllab
# array([0, 1, 0, 1, 0, 0, 1...
out.test_cllab
# array([0, 0, 0, 0, 1...
out.train_acc
# 1.0
out.test_acc
# 1.0
Attribute cv_results_ of FindBestClustCV returns a dataframe with training and validation
misclassification errors.
findbestclust.cv_results_
To visualize performance metrics during cross-validation, i.e., training stability and validation normalized stability with confidence intervals:
from reval.visualization import plot_metrics
plot_metrics(metrics, title="Reval metrics")
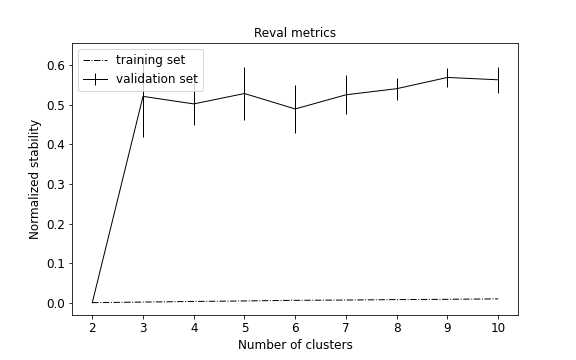
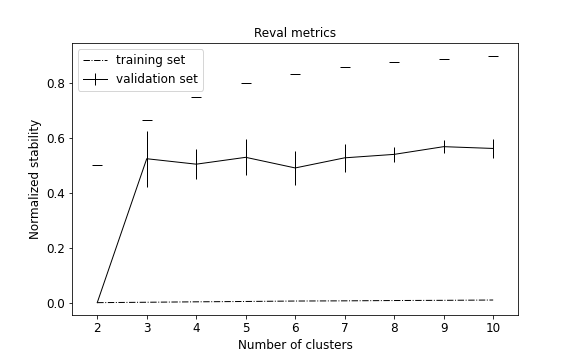
The plot can be customized and also show the normalized stability of a random classifier for each number of clusters to evaluate the model performance.
Classifier/clustering selection¶
Let us now suppose that we are not sure which combination of clustering and classifier to use
for the blobs dataset. We might want to try both hierarchical clustering and k-means and KNN and
logistic regression. We import the libraries we have not imported before including the
SCParamSelection from the param_selection.py module.
from sklearn.linear_model import LogisticRegression
from sklearn.cluster import KMeans
from reval.param_selection import SCParamSelection
We initialize the parameter selection class with a dictionary that includes the classification and clustering algorithms we want to run and we initialize a 10x2 repeated cross validation with 10 runs of random labeling. We set the number of parallel processes to 7 to speed up computations.
sc_params = {'s': [LogisticRegression(max_iter=1000), KNeighborsClassifier()],
'c': [AgglomerativeClustering(), KMeans()]}
scparsel = SCParamSelection(sc_params, cv=2, nrand=10, n_jobs=7,
iter_cv=10, clust_range=list(range(2, 11)),
strat=y_tr)
scparsel.fit(X_tr, nclass=2)
In this case we knew the true number of clusters a priori, so we passed it to the fit() method in
order to prioritize the parameter combinations that select the true number of clusters, along with the
combinations with global minimum stability. As a result, four different combinations are run and all of
them selected two as the best number of clusters with minimum stability.
Parameter selection¶
Let us now settle with hierarchical clustering and KNN and suppose we want to try different number of
neighbors for KNN, i.e., 5 and 15, and different methods for hierarchical clustering,
i.e., Ward and single-linkage. We can then use the ParamSelection as follows:
from reval.param_selection import ParamSelection
params = {'s': {'n_neighbors': [5, 15]},
'c': {'linkage': ['ward', 'single']}}
parsel = ParamSelection(params, cv=2, s=KNeighborsClassifier(), c=AgglomerativeClustering(),
nrand=10,
n_jobs=7,
iter_cv=10,
strat=y_tr, clust_range=list(range(2, 11)))
parsel.fit(X_tr, nclass=2)
Also in this case we run four different hyperparameter combinations which all report 2 as the best number of clusters with minimum stability.